 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold05987-augustus-gene-0.18-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 319aa MW: 35942.1 Da PI: 6.1693 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 46.8 | 6.7e-15 | 22 | 69 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEd++l ++ G g+W+ +ar g+ R++k+c++rw +yl
maker-scaffold05987-augustus-gene-0.18-mRNA-1 22 KGLWSPEEDDKLMNYMMNNGQGCWSDVARNAGLQRCGKSCRLRWINYL 69
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 47.2 | 5.1e-15 | 75 | 118 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ +++++ +lG++ W+ Ia++++ gRt++++k++w++
maker-scaffold05987-augustus-gene-0.18-mRNA-1 75 RGAFSPQEEAHIIHLHSLLGNR-WSQIAARLP-GRTDNEIKNFWNS 118
89********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 9.1E-24 | 15 | 72 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.138 | 17 | 69 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.52E-27 | 20 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-10 | 21 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.4E-14 | 22 | 69 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.73E-9 | 25 | 69 | No hit | No description |
| PROSITE profile | PS51294 | 24.149 | 70 | 124 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-25 | 73 | 125 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-13 | 74 | 122 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.1E-14 | 75 | 118 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.04E-9 | 77 | 117 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MRKPELSSVG KNNHVNNNKL RKGLWSPEED DKLMNYMMNN GQGCWSDVAR NAGLQRCGKS 60 CRLRWINYLR PDLKRGAFSP QEEAHIIHLH SLLGNRWSQI AARLPGRTDN EIKNFWNSTI 120 KKRLKNFSST PSPNTSDSSS PKDVMGGLIS MQEQGIMPLY LDSSSSTTNS MQAMTLNHMI 180 DPLPMIHHGL NMSNNASGYF NAAPQYMTQI GVNGDSFYGG NGVFGGADMG LENELFVPPL 240 ESISIEENDK TENMYNKRTM NNNTSNNLNN ISNCSNNNNR AENLARVGNY WEGEELRMGE 300 WDFEELMDVP SFPLLDFSN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 4e-26 | 15 | 124 | 20 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00599 | PBM | Transfer from AT5G12870 | Download |
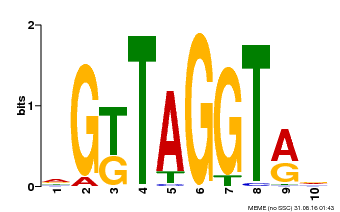
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023884289.1 | 0.0 | transcription factor MYB46-like | ||||
| TrEMBL | A0A2N9IV16 | 1e-175 | A0A2N9IV16_FAGSY; Uncharacterized protein | ||||
| STRING | cassava4.1_011597m | 1e-142 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF774 | 34 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G12870.1 | 5e-71 | myb domain protein 46 | ||||



