 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold06320-augustus-gene-0.9-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 413aa MW: 44691.2 Da PI: 6.3217 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39 | 1.4e-12 | 347 | 392 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L++l+P+ +k + a++L +AveYIk Lq
maker-scaffold06320-augustus-gene-0.9-mRNA-1 347 SIAERVRRTRISERMRKLQDLVPNM----DKQTNTADMLDLAVEYIKGLQ 392
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.919 | 341 | 391 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.94E-16 | 344 | 404 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.10E-12 | 345 | 396 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.8E-15 | 346 | 404 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-9 | 347 | 392 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.0E-14 | 347 | 397 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 413 aa Download sequence Send to blast |
MESDLQQHHH QNQMNSGLTR YRSAPSSYFA NIIDREFCEE FFNRPPSPET ERIFARFIAG 60 NGGVGGNGGT EATNTSEVVA TEASNQQTQF MASMNNEAAG VLQQQQSNYG SASQNFYQGP 120 PRPPLPNSSV EGSYSMGLDR LPQMKNGGGG GGGGGNNSSL IRHSSSPAGL FANINVGEGY 180 GTMRGVGNFG AGNSTSEEAS FSSASRLKNY SSGPPSSSGL MSPIDEIEDK NVVVVNNPES 240 GGFGEGRSSN YVTGFPLGSW DDSAMISDNI TGLKRLRDDD DVKSFSNINV SETQNVEAGN 300 RPPPLAHHLS LPKTSSEMAA IEKFLQFQDS VPCKIRAKRG CATHPRSIAE RVRRTRISER 360 MRKLQDLVPN MDKQTNTADM LDLAVEYIKG LQQEVETLTD KRAKCTCSHK QQQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
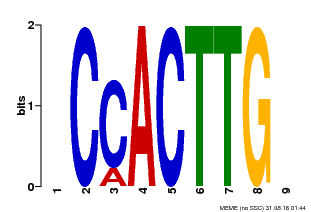
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023916484.1 | 0.0 | transcription factor bHLH130-like isoform X1 | ||||
| Refseq | XP_023916485.1 | 0.0 | transcription factor bHLH130-like isoform X1 | ||||
| TrEMBL | A0A2N9J1C8 | 0.0 | A0A2N9J1C8_FAGSY; Uncharacterized protein | ||||
| STRING | XP_008352410.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4185 | 34 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51140.1 | 3e-78 | bHLH family protein | ||||



