 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold06436-augustus-gene-0.12-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 518aa MW: 56581.5 Da PI: 4.829 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48 | 2.8e-15 | 31 | 78 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed +l ++vk++G g+W+++ + g+ R++k+c++rw ++l
maker-scaffold06436-augustus-gene-0.12-mRNA-1 31 KGPWTAAEDSILMEYVKKFGEGNWNAVQKNCGLMRCGKSCRLRWANHL 78
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 45.7 | 1.5e-14 | 84 | 127 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g+++ eE+ ++++++++lG++ W++ a+ ++ gRt++++k++w++
maker-scaffold06436-augustus-gene-0.12-mRNA-1 84 KGSFSGEEERIIIELHAKLGNK-WARMAAQLP-GRTDNEIKNYWNT 127
68999*****************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.256 | 26 | 78 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.13E-29 | 28 | 125 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.0E-12 | 30 | 80 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.4E-14 | 31 | 78 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-23 | 32 | 85 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.37E-10 | 33 | 78 | No hit | No description |
| PROSITE profile | PS51294 | 24.714 | 79 | 133 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.2E-15 | 83 | 131 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-13 | 84 | 127 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-24 | 86 | 132 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.58E-11 | 86 | 127 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 518 aa Download sequence Send to blast |
MISKSVEGAQ NQEAMAAAKG EAELSRAGLK KGPWTAAEDS ILMEYVKKFG EGNWNAVQKN 60 CGLMRCGKSC RLRWANHLRP NLKKGSFSGE EERIIIELHA KLGNKWARMA AQLPGRTDNE 120 IKNYWNTRMK RRQRAGLPLY PQEVQQEAAV FHLLQQQNHQ KQPHSSSVPF SAFLPSSQPR 180 KLNYNPSLSV FNTMNFSSPT NPIQNQVTPD FYSNPSNQFK FFSENNNNPA YALPLSPVSP 240 FGSSPSPIFN QNIAPQPLSA PFVEYNSDSF GSSQYNRSPA VMGPAPYEPF GLVQGLETEL 300 PSNQTPLNST THTSSTSSGD ENPLGASSSA NDFEIENPLS QSPDNSGLLD ALLVESQTLC 360 EKSKGKEVSA AVASDKGKGV MVEASTEEED PMNVDSVFKT SGETSAENHW DDLSSSQSQS 420 QSQSSIGMKP AEDPLEEMNS MDDDLASLLT NFPSAMPTPE WYRGGHHLSN GQSSGVESDN 480 VRLDTQQSAS PALVTNTTEP ELESGLSPCY WKNLPGIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-32 | 29 | 132 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
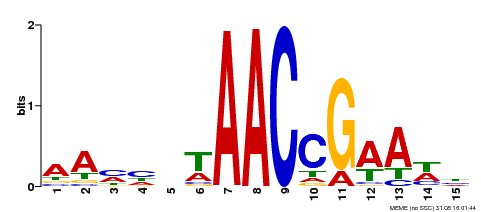
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023886859.1 | 0.0 | transcription factor MYB101-like | ||||
| TrEMBL | A0A2N9I547 | 0.0 | A0A2N9I547_FAGSY; Uncharacterized protein | ||||
| STRING | EOY02206 | 1e-140 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9949 | 32 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.1 | 2e-81 | myb domain protein 101 | ||||



