 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold13322-augustus-gene-0.2-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 532aa MW: 58901.4 Da PI: 4.9792 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 84.2 | 1e-26 | 181 | 246 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekn 65
r+++sL+llt+kf++ll+++e+g+++ln++a++L +v ++RRiYDi+NVLe+++liekk kn
maker-scaffold13322-augustus-gene-0.2-mRNA-1 181 RYDSSLGLLTKKFINLLKQAEDGNLDLNKAAETL---EV--QKRRIYDITNVLEGIGLIEKKLKN 240
6899******************************...99..************************ PP
E2F_TDP 66 eirwkg 71
+i+wkg
maker-scaffold13322-augustus-gene-0.2-mRNA-1 241 RIHWKG 246
****98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.4E-28 | 179 | 248 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 2.8E-34 | 181 | 246 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 4.79E-17 | 181 | 244 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF02319 | 1.6E-24 | 183 | 246 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF144074 | 3.4E-31 | 286 | 388 | No hit | No description |
| Pfam | PF16421 | 3.3E-32 | 287 | 386 | IPR032198 | E2F transcription factor, CC-MB domain |
| CDD | cd14660 | 2.22E-45 | 287 | 388 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051446 | Biological Process | positive regulation of meiotic cell cycle | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 532 aa Download sequence Send to blast |
MSGGARAPNR PAPPPPTVAS QILPPPPPMK RNLAVLASTK PPFVPPDDYH RFSSVVADRE 60 AEALVVRSPI DSDDCVLVVW HEASGCLFVL AAGDSQLLKR KNGTDDNEVQ SIDWTNSPGY 120 TSVVNSPLKT PVSAKGGRIY NRSKGTKGNR SGPQTPVSNA GETTFPHSLR SPSPLTPGSC 180 RYDSSLGLLT KKFINLLKQA EDGNLDLNKA AETLEVQKRR IYDITNVLEG IGLIEKKLKN 240 RIHWKGLDAL RPGEVEDDAS VLQGEKKGKK DKQFNNFSWK MKICLFLQAE VENLTNQEQR 300 LDDQIREMQE RLRDLSEDEN KKKWLFVTKE DIKGLPCLQN ETLLAIKAPH GTTLEVPDPD 360 EAVDYPQRRY RVILRSAMGP IDVYLVSQFE EKFEEMNSVE PPVSFPMVSS SGSNDNPATD 420 EVVNANSTMK DVMPQAQHSH QMCSDLNASQ EFPGGMMKIV PSDVDNDADY WLLSDAEVSI 480 TDMWKTDAGI EWSGMDMLHP DFGIPDVSTP RSQTPLSGMA EVPSTAFDST RR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 8e-24 | 177 | 247 | 2 | 74 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The binding of retinoblastoma-related proteins represses transactivation. Regulates gene expression both positively and negatively. Activates the expression of E2FB. Involved in the control of cell-cycle progression from G1 to S phase. Stimulates cell proliferation and delays differentiation. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11862494, ECO:0000269|PubMed:11889041, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12913157, ECO:0000269|PubMed:15377755, ECO:0000269|PubMed:16514015, ECO:0000269|PubMed:19662336}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00293 | DAP | Transfer from AT2G36010 | Download |
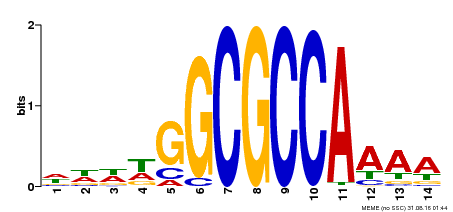
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023886542.1 | 0.0 | transcription factor E2FA isoform X1 | ||||
| Swissprot | Q9FNY0 | 1e-159 | E2FA_ARATH; Transcription factor E2FA | ||||
| TrEMBL | A0A2N9ILM9 | 0.0 | A0A2N9ILM9_FAGSY; Uncharacterized protein | ||||
| STRING | VIT_08s0007g02510.t01 | 0.0 | (Vitis vinifera) | ||||
| STRING | EMJ05533 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1947 | 34 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G36010.3 | 1e-161 | E2F transcription factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



