 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | model.Picochlorum_contig_155.g704.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Picochlorum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 411aa MW: 45955.4 Da PI: 6.6855 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.9 | 3.9e-32 | 139 | 193 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+rlrWtpeLH+rFv av++LGG+ekAtPk il+lm+v++Lt++h+kSHLQkYRl
model.Picochlorum_contig_155.g704.t1 139 KARLRWTPELHQRFVGAVHSLGGPEKATPKGILNLMNVEDLTIYHIKSHLQKYRL 193
68****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.863 | 136 | 196 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.7E-31 | 136 | 195 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.15E-17 | 137 | 196 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.2E-25 | 139 | 193 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.2E-8 | 141 | 192 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.7E-15 | 269 | 312 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 411 aa Download sequence Send to blast |
MKTPGEDNPD HQNDAQLLDL EYWPQLDLAP ELFNLEWIGG EQEHHHINGP LPGSPHQGKL 60 GISLDGSRLV SRGSPKEHND EVKRDVEPMH VVDDIAIFQD VDRGMDLQLD CGEDRSFMEN 120 MDSGLEERKQ RRRVQSSSKA RLRWTPELHQ RFVGAVHSLG GPEKATPKGI LNLMNVEDLT 180 IYHIKSHLQK YRLNIKVPGE EKHRREEEKE SVQKSKRRIS RKKSTQPGLD HQGSLDREGS 240 LPHKASTSVQ QTTSIDSLTQ RGSVEEHRIQ LERALLVQME MQKKLHEQLE AQRKLQMNLE 300 QHGRYISSLL ESTGLAGTVH GSQIDENGNK KAVPGDDFGN LKTSLNSMGM SAEKLKNLYI 360 KNIGSNISAA PSSDHFSHSI LMNGELAGAP GAQKGGDYYI QEEPGGRDGK * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 9e-22 | 139 | 192 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-22 | 139 | 192 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-22 | 139 | 192 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-22 | 139 | 192 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
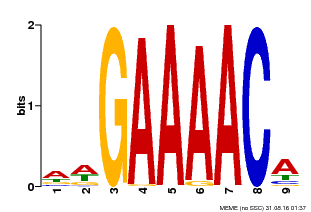
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP111 | 16 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G13640.1 | 6e-31 | G2-like family protein | ||||



