 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | model.Picochlorum_contig_16.g153.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Picochlorum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 245aa MW: 27100.5 Da PI: 6.8107 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 54.3 | 2.9e-17 | 175 | 236 | 1 | 62 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
ek++k+ rrkq+NRe+ArrsR+RK+ae+e+L+++ +L+ae ++L+ke elk ++ l+s+
model.Picochlorum_contig_16.g153.t1 175 EKDIKKLRRKQSNRESARRSRLRKQAECEHLQNENFTLKAEVDRLRKEQIELKTTITVLQSQ 236
899***************************************************99988876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.2E-13 | 173 | 240 | No hit | No description |
| SMART | SM00338 | 1.7E-13 | 175 | 239 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.0E-14 | 175 | 236 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.013 | 177 | 240 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.19E-10 | 178 | 237 | No hit | No description |
| CDD | cd14702 | 9.07E-15 | 180 | 237 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 182 | 197 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 245 aa Download sequence Send to blast |
MDGEGPRNAV EIPDVAQASD EMRKHWEQIQ ENMLKSFPNT LPFGNGAMAA PFPMPPALMP 60 LAMMQMSNLA SLYSSGLVGG EVPSVEQKTQ TENATNNNDK TLSGLNLLAS TVGQPTTNNN 120 FPGINSPLVS EKYSGDAKTG KSDSQQAKRS RSLSLTQDQD AEIYLDEATI AGMDEKDIKK 180 LRRKQSNRES ARRSRLRKQA ECEHLQNENF TLKAEVDRLR KEQIELKTTI TVLQSQLLEL 240 RKHA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 179 | 184 | KLRRKQ |
| 2 | 191 | 197 | RRSRLRK |
| 3 | 191 | 198 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
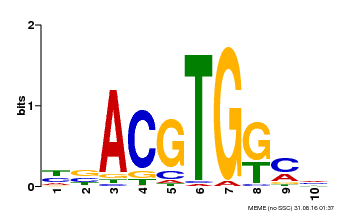
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP4426 | 9 | 11 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.1 | 1e-16 | G-box binding factor 1 | ||||



