 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | model.Picochlorum_contig_196.g913.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Picochlorum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 277aa MW: 30462.6 Da PI: 9.9913 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45.7 | 1.4e-14 | 207 | 259 | 2 | 54 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkk 54
+++k r+++NRe+A rsR+++++++ eLe kvk+L+ Nk+L++++ +l k
model.Picochlorum_contig_196.g913.t1 207 RSVKSDERAMRNRESAARSRAKRLEYTHELEVKVKTLKDVNKELREKIIKLAK 259
6788999*****************************************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00170 | 2.5E-12 | 207 | 259 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.795 | 208 | 259 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 1.4E-6 | 209 | 270 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.81E-9 | 210 | 257 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 1.2E-12 | 212 | 258 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 214 | 228 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 277 aa Download sequence Send to blast |
MQQNVDWDAL LRSPATETVQ HDHLSDIQLG QLAEGIENLE AELPPGVVEG SSLAPGLRDV 60 RLAEVIGPVI QGQETSTLNV AQCLDLWSRG LNASGPSHAG VVPQQCYPPM AAHAGQHVIQ 120 GSVLNIQLAP LHNRIDEEKE MAAMNRAERQ RKRKEKQEES VRRKMSAHAS GSEMVPIPTG 180 LSSSQITVGA ENQSKEGIKT NGPRRRRSVK SDERAMRNRE SAARSRAKRL EYTHELEVKV 240 KTLKDVNKEL REKIIKLAKA PADPNGALRR TRTMPL* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 150 | 155 | RKRKEK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00409 | DAP | Transfer from AT3G56850 | Download |
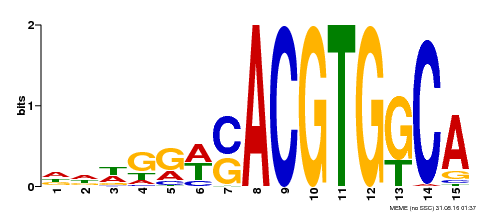
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP9391 | 4 | 4 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56850.1 | 4e-12 | ABA-responsive element binding protein 3 | ||||



