 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | model.Picochlorum_contig_84.g402.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Picochlorum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 215aa MW: 24572.8 Da PI: 10.4111 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.1 | 2.3e-14 | 132 | 176 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + +++G+g+W++I+r + +Rt+ q+ s+ qky
model.Picochlorum_contig_84.g402.t1 132 PWTEEEHRLFLMGLAKYGKGDWRSISRNFVITRTPTQVASHAQKY 176
7*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 6.914 | 38 | 92 | IPR017884 | SANT domain |
| SMART | SM00717 | 0.82 | 39 | 90 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 7.59E-6 | 40 | 94 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 0.00167 | 43 | 87 | No hit | No description |
| PROSITE profile | PS51294 | 18.835 | 125 | 181 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.9E-17 | 126 | 182 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.6E-17 | 129 | 179 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.1E-12 | 129 | 179 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-12 | 130 | 175 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.5E-13 | 132 | 176 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.25E-10 | 132 | 177 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 215 aa Download sequence Send to blast |
MVSGTRSAGN GTKQQQQKKD AHDGSHAKLA EGKADHAVAK GGWSEEEDRV FENSLAQYWD 60 FPDRFEKCAS MLSRKNLTDV IARFKELDED IRNIEMGRVQ MPAYPVPGEA LSISQLQKKV 120 KSQDTERRKG IPWTEEEHRL FLMGLAKYGK GDWRSISRNF VITRTPTQVA SHAQKYFIRL 180 NSQNKKDKRR ASIHDITSVH PEYKKKPKKS TKKE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00568 | DAP | Transfer from AT5G58900 | Download |
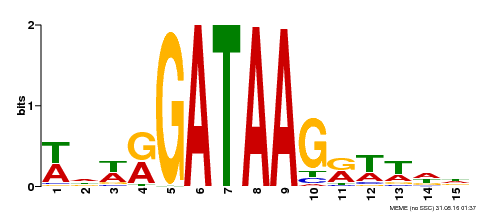
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005844178.1 | 1e-87 | expressed protein | ||||
| Swissprot | Q8S9H7 | 1e-51 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | E1ZQ14 | 3e-86 | E1ZQ14_CHLVA; Expressed protein | ||||
| STRING | XP_005844178.1 | 5e-87 | (Chlorella variabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP322 | 15 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58900.1 | 9e-57 | Homeodomain-like transcriptional regulator | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



