 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | mrna08853.1-v1.0-hybrid | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 303aa MW: 34215.5 Da PI: 8.7256 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.5 | 5.5e-19 | 127 | 181 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ +++k+q +Lee F+++++++ +++ LAk+lgL rqV vWFqNrRa+ k
mrna08853.1-v1.0-hybrid 127 RKKLRLSKDQSAILEESFKEHNTLNPKQKLALAKQLGLRPRQVEVWFQNRRARTK 181
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.6 | 1.2e-41 | 127 | 216 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekevee 86
+kk+rlsk+q+++LEesF+e+++L+p++K +la++Lgl+prqv+vWFqnrRARtk+kq+E+d+e+Lkr++++l+een+rL+kev+e
mrna08853.1-v1.0-hybrid 127 RKKLRLSKDQSAILEESFKEHNTLNPKQKLALAKQLGLRPRQVEVWFQNRRARTKLKQTEVDCEFLKRCCENLTEENRRLQKEVQE 212
69************************************************************************************ PP
HD-ZIP_I/II 87 Lreel 91
Lr +l
mrna08853.1-v1.0-hybrid 213 LR-AL 216
*9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04618 | 8.0E-16 | 47 | 101 | IPR006712 | HD-ZIP protein, N-terminal |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-18 | 113 | 177 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.0E-18 | 117 | 184 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.443 | 123 | 183 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-16 | 125 | 187 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.0E-16 | 127 | 181 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.38E-15 | 127 | 184 | No hit | No description |
| PRINTS | PR00031 | 2.1E-5 | 154 | 163 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 158 | 181 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 2.1E-5 | 163 | 179 | IPR000047 | Helix-turn-helix motif |
| SMART | SM00340 | 4.2E-27 | 183 | 226 | IPR003106 | Leucine zipper, homeobox-associated |
| Pfam | PF02183 | 2.3E-11 | 183 | 217 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008283 | Biological Process | cell proliferation | ||||
| GO:0009641 | Biological Process | shade avoidance | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010016 | Biological Process | shoot system morphogenesis | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MSSNDCDEEG VGKWRDKMTR TTARCGVFVW GPPMWGGRAW ESTCSEGPRR TYRSSDTCRG 60 PARSFLRGID VNRLPSTGDC EDVSSPNSTV SSVSGKRSER EANGEELDLE RDGSRGMSDE 120 EDGETSRKKL RLSKDQSAIL EESFKEHNTL NPKQKLALAK QLGLRPRQVE VWFQNRRART 180 KLKQTEVDCE FLKRCCENLT EENRRLQKEV QELRALKLSP QFYMQMTPPT TLTMCPSCER 240 VAVPPNSTSP SATVDPRAHH HHHHPMVPVH HPRAIPVAGP WAAAAPVLQQ QRQFEPFRTA 300 RP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 125 | 131 | SRKKLRL |
| 2 | 175 | 183 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the negative regulation of cell elongation and specific cell proliferation processes such as lateral root formation and secondary growth of the vascular system. Acts as mediator of the red/far-red light effects on leaf cell expansion in the shading response. Binds to the DNA sequence 5'-CAAT[GC]ATTG-3'. Negatively regulates its own expression. {ECO:0000269|PubMed:10477292, ECO:0000269|PubMed:11260495, ECO:0000269|PubMed:8449400}. | |||||
| UniProt | Probable transcription factor that plays a role in auxin-mediated morphogenesis. Negatively regulates lateral root elongation. {ECO:0000269|PubMed:12492842}. | |||||
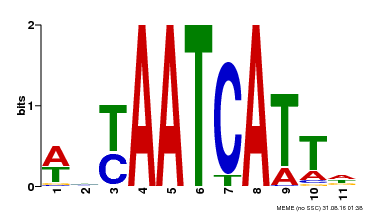
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00550 | DAP | Transfer from AT5G47370 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | mrna08853.1-v1.0-hybrid |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By indole-3-acetic acid (IAA). {ECO:0000269|PubMed:12492842}. | |||||
| UniProt | INDUCTION: Rapidly and strongly induced by lowering the ratio of red to far-red light. {ECO:0000269|PubMed:8106086}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004301172.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HAT4-like | ||||
| Swissprot | P46601 | 4e-92 | HAT2_ARATH; Homeobox-leucine zipper protein HAT2 | ||||
| Swissprot | Q05466 | 3e-92 | HAT4_ARATH; Homeobox-leucine zipper protein HAT4 | ||||
| TrEMBL | A0A2P6P8P0 | 1e-154 | A0A2P6P8P0_ROSCH; Putative transcription factor HD-ZIP family | ||||
| STRING | XP_004301172.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1239 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16780.1 | 1e-85 | homeobox protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | mrna08853.1-v1.0-hybrid |



