 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | mrna16296.1-v1.0-hybrid | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1193aa MW: 134132 Da PI: 6.0799 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 74.9 | 1.1e-23 | 89 | 166 | 2 | 81 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltl...elktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvgg.vevl 81
l+ k+rwl++ ei++iL+n++k+++++ +++ +p g+l ++ryfrkDG++w+kkkdgktv+E+he+LK +vl
mrna16296.1-v1.0-hybrid 89 LEAKHRWLRPAEICEILQNYKKFHISTepaSTPPEPARGHL------VLRYFRKDGHNWRKKKDGKTVKEAHERLKKTDhFQVL 166
6679*********************9944444555566666......569*************************853325555 PP
| |||||||
| 2 | CG-1 | 59 | 9.1e-19 | 205 | 254 | 69 | 118 |
CG-1 69 dhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
+ L+ g+v+vl+cyYah+e+n++fqrr+yw+Lee+l++ivlvhy+evk
mrna16296.1-v1.0-hybrid 205 TDIGLRAGSVDVLHCYYAHGEDNENFQRRSYWMLEEDLSHIVLVHYREVK 254
55679999***************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 62.426 | 84 | 259 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.0E-45 | 87 | 254 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 7.9E-19 | 90 | 160 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.2E-15 | 204 | 252 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.8E-6 | 617 | 703 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.26E-18 | 618 | 703 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 7.8E-7 | 618 | 702 | IPR002909 | IPT domain |
| PROSITE profile | PS50297 | 18.139 | 783 | 922 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 9.48E-17 | 797 | 912 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 8.97E-13 | 798 | 910 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-17 | 801 | 914 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.217 | 851 | 883 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.032 | 851 | 880 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 260 | 890 | 919 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.33E-7 | 1009 | 1069 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 23 | 1018 | 1040 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.089 | 1020 | 1048 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.18 | 1021 | 1039 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0017 | 1041 | 1063 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.597 | 1042 | 1066 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.3E-4 | 1044 | 1063 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1193 aa Download sequence Send to blast |
MKYRLDLGVQ SNYILYAIGI NVSHSWKKTK ASDIQLSSPC VFPKMFRLRD FLFGEDPYGY 60 IRRGSALGFC IAEAKTKVCS ILDIQQILLE AKHRWLRPAE ICEILQNYKK FHISTEPAST 120 PPEPARGHLV LRYFRKDGHN WRKKKDGKTV KEAHERLKKT DHFQVLMEIW VNLRLRLVCR 180 LIKANELSSL SFLLASAELQ LNLETDIGLR AGSVDVLHCY YAHGEDNENF QRRSYWMLEE 240 DLSHIVLVHY REVKGNRTNF NHVKETEGVA YSNGAEQSAR QSEMENSVSS SFNPSSYQMH 300 SQTTEATSLS SAQASEFEDA ESAFYNQASS RLQPMAEKIN SEFADAYYPT FSNDFQEKLS 360 TIPGVDFSSL SQAYKGEDSI HAGITHEPRK DRDFALWDDM ENSATGVQSF QPSFSATHSD 420 TMGSFPKQEI ETIGHLYTDS FDKRLVYGME NRPKVQQSWQ TSEGSSNWPM DQSIQSHAQY 480 NVTSKLHDGA DATDLLKSLG PFLMDSDKQN DLQFHLSNTD SISKRNDIIE GKADYPSAIK 540 PLLDGAFGDG LKKLDSFNRW MSKELEDVDE PQMQSSSGAY WETVESENEV DESSVPLQVR 600 LDSYMLGPSL SHDQLFSIVD FSPSWAYENS EIKVLITGRF LKSQHAESCK WSCMFGEVEV 660 PAEVIADGVL RCYTPIHKAG RVPFYVTCSN RLACSEVREF EYRVAETQDV DCKDYYSDFS 720 NETLSMRFGN FLTLSSTSPN CDPASIAENS EVNSKITSLL KNDNDEWDKM LQLTSDEDFS 780 LKRVEEQLHQ QLLKEKLHAW LLQKLAAGGK GPNVLDEGGQ GVLHFGAALG YDWVLLPTIT 840 AGVSVNFRDV NGWTALHWAA FCGRERTVAS LISLGAAPGA LTDPTAKYPS GETPADLASE 900 QGHKGIAGYL AESALSKHLE SLNLDIKDGN SAEISGAKAV SGSSRDGELT DGLSLRDSLT 960 AVCNATQAAA RIHQVFRVQS FQRKQLKEYG GDKFGISNER ALSLIAVKSH KAGKRDEHVD 1020 AAAVRIQNKF RSWKGRKDFL IIRQRIVKIQ AHVRGHQVRK NYKKIVWTVG IVEKIILRWR 1080 RKGSGLRGFK PEPLTEGPSM QVSSTKEDDD DVLKEGRKQT EERMQKALAR VKSMAQYPEA 1140 RDQYRRLLNV VTEIQETKVL NSSEGTSAYM DDDLIDIEAL FDDDVFMPTA TS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cxk_A | 3e-14 | 618 | 703 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_B | 3e-14 | 618 | 703 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_C | 3e-14 | 618 | 703 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_D | 3e-14 | 618 | 703 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_E | 3e-14 | 618 | 703 | 9 | 89 | calmodulin binding transcription activator 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
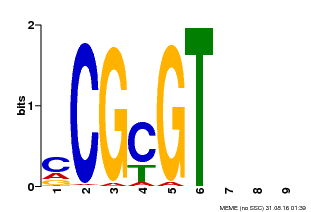
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | mrna16296.1-v1.0-hybrid |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM434540 | 4e-36 | AM434540.2 Vitis vinifera contig VV78X210461.8, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004288193.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3-like | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A2P6RQI8 | 0.0 | A0A2P6RQI8_ROSCH; Putative transcription factor CG1-CAMTA family | ||||
| STRING | XP_004288193.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8284 | 27 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | mrna16296.1-v1.0-hybrid |



