 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | mrna19130.1-v1.0-hybrid | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 830aa MW: 89980.7 Da PI: 6.3188 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.6 | 3.2e-21 | 131 | 186 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+++l+L++rqVk+WFqNrR+++k
mrna19130.1-v1.0-hybrid 131 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSRRLNLETRQVKFWFQNRRTQMK 186
688999***********************************************999 PP
| |||||||
| 2 | START | 208.8 | 2e-65 | 335 | 557 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE..EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S.. CS
START 1 elaeeaaqelvkkalaeepgWvkss..esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla.. 77
ela++a++elvk+a+ +ep+W+++ e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++
mrna19130.1-v1.0-hybrid 335 ELALAAMDELVKLAQTDEPLWSLEGgrEILNHEEYMRSFTPCIGlkpngFVTEASRETGMVIINSLALVETLMDSN-RWLEMFPcm 419
5899********************999*************9999999*9***************************.********* PP
..EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEE CS
START 78 ..kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgili 154
+ +t++vissg galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvSvd ++++ + ++ +++lpSg+++
mrna19130.1-v1.0-hybrid 420 iaRTSTTDVISSGmggtrnGALQLMHAELQVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSVDTIRDNSGAPTFANCRRLPSGCVV 505
*********************************************************************9**************** PP
EEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 155 epksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++ng+skvtwveh++++++++h l+r+l++sg+ +ga++wvatlqrqc++
mrna19130.1-v1.0-hybrid 506 QDMPNGYSKVTWVEHAEYDESQVHHLYRPLLSSGMGFGAQRWVATLQRQCQC 557
**************************************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-22 | 117 | 188 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.17E-21 | 117 | 188 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.058 | 128 | 188 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.6E-19 | 129 | 192 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 8.2E-19 | 131 | 186 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.20E-19 | 131 | 188 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 163 | 186 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 43.593 | 326 | 560 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 3.57E-33 | 330 | 557 | No hit | No description |
| CDD | cd08875 | 1.01E-124 | 332 | 556 | No hit | No description |
| SMART | SM00234 | 4.8E-49 | 335 | 557 | IPR002913 | START domain |
| Pfam | PF01852 | 2.0E-57 | 335 | 557 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.26E-23 | 586 | 756 | No hit | No description |
| SuperFamily | SSF55961 | 6.26E-23 | 783 | 823 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 830 aa Download sequence Send to blast |
MSFGGFLDNS TGSSGGARIV ADIPYNHHPH HNANHTSMPS SAIAQPRLVT QSLTKSMFNN 60 SPGLSLALQT NADGGGDAAR MAENFEGNNN VGGRRSREEE NEISRSGSDN MDGAGSGDEG 120 DAADNSNPRK KKRYHRHTPQ QIQELEALFK ECPHPDEKQR LELSRRLNLE TRQVKFWFQN 180 RRTQMKTQLE RHENSLLRQE NDKLRAENMS IRDAMRNPIC TNCGGPAMIG DISIEEQHLR 240 IDNARLKDEL DRVCALAGKF LGRPISSLGP SMGPPLPSSA LELGVGNNGF GGMSSVSTSM 300 PLGPDFGAGL GGGMPLVAHT RPVAGGLDER TMFLELALAA MDELVKLAQT DEPLWSLEGG 360 REILNHEEYM RSFTPCIGLK PNGFVTEASR ETGMVIINSL ALVETLMDSN RWLEMFPCMI 420 ARTSTTDVIS SGMGGTRNGA LQLMHAELQV LSPLVPVREV NFLRFCKQHA EGVWAVVDVS 480 VDTIRDNSGA PTFANCRRLP SGCVVQDMPN GYSKVTWVEH AEYDESQVHH LYRPLLSSGM 540 GFGAQRWVAT LQRQCQCLAI LMSSTVPARD HANTITQSGR KSMLKLAQRM TDNFCAGVCA 600 STVHKWNKLN AGNVDEDVRY MTRESMDDPG EPPGIVLSAA TSVWLPVSPQ RLFNFLRDER 660 LRSEWDILSN GGPMQEMAHI AKGQDQGNCV SLLRARAMNA NQNSMLILQE TCIDAAGSLV 720 VYAPVDIPAM HVVMNGGDSA YVALLPSGFA IVPDGPGSRG PGGAEGKAGQ GSSNGNGGEA 780 RVSGSLLTMT FQILVNSLPS AKLTVESVET VNNLISCTVQ KIKGALQCES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
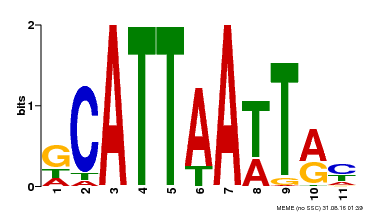
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | mrna19130.1-v1.0-hybrid |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004306832.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A2P6SGK8 | 0.0 | A0A2P6SGK8_ROSCH; Putative transcription factor & lipid binding Homobox-WOX family | ||||
| STRING | XP_004306832.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | mrna19130.1-v1.0-hybrid |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



