 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | mrna22961.1-v1.0-hybrid | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Rosoideae; Potentilleae; Fragariinae; Fragaria
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 442aa MW: 49138.3 Da PI: 7.6473 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.1 | 2e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l+++++++G g+W+++++ g+ R++k+c++rw +yl
mrna22961.1-v1.0-hybrid 14 KGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45 | 2.5e-14 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ l++++++ lG++ W+ Ia+ ++ gRt++++k+ w++
mrna22961.1-v1.0-hybrid 67 RGPFSQQEENLIIELHAVLGNR-WSQIAAQLP-GRTDNEIKNLWNS 110
89********************.*********.*********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-27 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.407 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.24E-30 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.2E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.61E-12 | 17 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.421 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.2E-12 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.4E-13 | 67 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.20E-9 | 69 | 109 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010214 | Biological Process | seed coat development | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009001 | anatomy | fruit | ||||
| PO:0007010 | developmental stage | whole plant fruit ripening stage | ||||
| PO:0007027 | developmental stage | whole plant fruit formation stage 70% to final size | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 442 aa Download sequence Send to blast |
MGRHSCCYKQ KLRKGLWSPE EDEKLLNYIT KHGHGCWSSV PKLAGLQRCG KSCRLRWINY 60 LRPDLKRGPF SQQEENLIIE LHAVLGNRWS QIAAQLPGRT DNEIKNLWNS CIKKKLRQRG 120 IDPNTHKPIS SETEQTDHSK EISQLSSPTN YKGNEQQKAS VGSNELNLVE AVANSKQVEH 180 HRYPVEVSSS SKVISNSNNT QDFFIDRAAC TSHEGSTTTN CRPSDFVGYF SFHHHHQFGS 240 SSSETMGLNA VNPNQTFSFL NQNSRSNSDP QMVSEFSTSM TTPPSLLPSM SSSSLFGTRV 300 KPSISLPSDN SSTVSWDQSS AGYSNNGGFD NTAAAFSWGL LPEVPVKSDD VEEINKWSEY 360 IHSPFLMGAN TMQNINQSSQ YNSTEIKPET QYFMTSNGAS SASAATTSWP HHHQQANNLQ 420 PSSEMYTKDL QRLAVAFGQT L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 6e-28 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
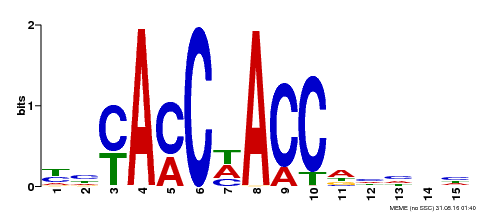
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00134 | DAP | Transfer from AT1G09540 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | mrna22961.1-v1.0-hybrid |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004302323.1 | 0.0 | PREDICTED: transcription factor MYB86 | ||||
| TrEMBL | A0A2P6R7X1 | 0.0 | A0A2P6R7X1_ROSCH; Putative transcription factor MYB-HB-like family | ||||
| STRING | XP_004302323.1 | 0.0 | (Fragaria vesca) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1083 | 33 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G57560.1 | 4e-88 | myb domain protein 50 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | mrna22961.1-v1.0-hybrid |



