 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g001365m | ||||||||
| Common Name | CISIN_1g001365mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1093aa MW: 122008 Da PI: 5.8795 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180 | 2.9e-56 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenpt 93
++ ++rwl++ ei++iL n++k++++ e++++p+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+v+vl+cyYah+een++
orange1.1g001365m 20 IEAQHRWLRPAEICEILRNYTKFRIAPESPHTPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSVDVLHCYYAHGEENEN 111
5679**************************************************************************************** PP
CG-1 94 fqrrcywlLeeelekivlvhylevk 118
fqrr+yw+Leeel++ivlvhy+evk
orange1.1g001365m 112 FQRRSYWMLEEELSHIVLVHYREVK 136
**********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 83.036 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 4.7E-78 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.0E-49 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.0E-5 | 509 | 597 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 3.4E-5 | 510 | 595 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 3.92E-17 | 510 | 596 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 4.2E-16 | 697 | 806 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 6.49E-12 | 701 | 804 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 8.9E-16 | 703 | 808 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.99 | 712 | 806 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.0E-6 | 717 | 807 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.351 | 745 | 777 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.024 | 745 | 774 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 610 | 784 | 813 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 9.29E-8 | 915 | 970 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 17 | 919 | 941 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.067 | 921 | 940 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.126 | 921 | 949 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0033 | 942 | 964 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.091 | 943 | 967 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.0E-4 | 945 | 964 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1093 aa Download sequence Send to blast |
MADSRRFALG NQLDIEQILI EAQHRWLRPA EICEILRNYT KFRIAPESPH TPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSVDVLH CYYAHGEENE NFQRRSYWML 120 EEELSHIVLV HYREVKGNRT NFNRAKVAEG ATPYSQENEE TIPNSEVEGS QSSGFHPNSY 180 QMPSQTADTS LNSAQASEYE DAESVYNNQA SSRFHSFLDL QQPVAEKIDA GLADPYYPSS 240 LTNNYQGKFS VVPGADFISP AQTDKSRNSN DTGLTYEPQK NLDFPSWEDV LQNCSQGVGS 300 QPEALGDIPN QGYDILGEPF TNSFGERKEF GSHLQTRGEW QASRNDSSHL SNWPMDQKVY 360 LDSAHDLTSQ SCEQGAAHDG LLDSLRPPHA HPNMENDVHE QLPNAEHGHL LKSDPESSLT 420 IDGKSFYSSA IKQHLIDGST EGLKKLDSFN RWMSKELGDV KESNMQSSSG AYWETVESEN 480 GVDDSGVSPQ ARLDTYMMSP SLSQDQLYSI IDFSPNWAYV SSEVKVLITG RFLMSQQEAE 540 NCKWSCMFGE IEVPAEIVAG GVLRCHTSSQ KVGRVPFYVT CSNRLSCSEV REFEYRASHI 600 PDVDVADNCG DITSENLRMQ FGKLLCLTSV STPNYDPSNL SDISQLNSKI SSLLKDENDD 660 WDLMLKLTAE EKFSSEEVKE KLVQKLLKEK LQVWLVQKAA EGGKGPCVLD HCGQGVLHFA 720 AALGYDWALE PTTVAGVNIN FRDVNGWTAL HWAAYCGRER TVASLIALGA APGALSDPTP 780 KYPSGRTPAD LASSIGHKGI AGYLAESDLS SALSAISLNK KDGDVAEVTG ATAVQTVPQR 840 CPTPVSDGDL PYGLSMKDSL AAVRNATQAA ARIHQVFRVQ SFQKKQLKEY GNDTFGISDE 900 RALSLVAVKT QKPGHHDEPV HAAATRIQNK FRSWKGRKDF LIIRKQIIKI QAYVRGHQVR 960 KNYKKIIWSV GIMEKIILRW RRRGSGLRGF KSETLTASSS MVATSAKEDD YDFLKEGRKQ 1020 KEERLQKALA RVKSMVQYPE ARDQYRRLLN VVNEIQETKA MALSNAEETA DFDDDLVDIE 1080 ALLDDTLMPN AS* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.17230 | 0.0 | callus | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, carpels, and siliques, but not in stigmas or other parts of the flower. {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
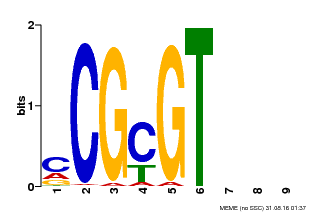
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU658749 | 8e-65 | EU658749.1 Sorindeia madagascariensis clone Sorin8 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006428204.1 | 0.0 | calmodulin-binding transcription activator 3 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A067F009 | 0.0 | A0A067F009_CITSI; Uncharacterized protein | ||||
| STRING | XP_006428204.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8601 | 25 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g001365m |



