 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g001406m | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1084aa MW: 121706 Da PI: 5.441 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188 | 9.2e-59 | 21 | 136 | 4 | 118 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktvrE+hekLKvg+v+vl+cyYah+e+n++f
orange1.1g001406m 21 EaQHRWLRPAEICEILCNYQKFHIASEPPSRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVREAHEKLKVGSVDVLHCYYAHGEDNENF 112
449***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrrcyw+Le++l +iv+vhylev+
orange1.1g001406m 113 QRRCYWMLEQDLMHIVFVHYLEVQ 136
*********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.221 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.0E-85 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.5E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.8E-5 | 482 | 583 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.01E-14 | 498 | 583 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 1.2E-16 | 680 | 793 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.37E-16 | 686 | 792 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.88E-13 | 687 | 790 | No hit | No description |
| PROSITE profile | PS50297 | 16.255 | 698 | 792 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0042 | 731 | 760 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.698 | 731 | 763 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.45E-8 | 905 | 956 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.099 | 905 | 927 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.151 | 906 | 935 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 9.2E-4 | 907 | 926 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.002 | 928 | 950 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.725 | 929 | 953 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.5E-4 | 931 | 950 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1084 aa Download sequence Send to blast |
MADRGSYALA PRLDMQQLQM EAQHRWLRPA EICEILCNYQ KFHIASEPPS RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVREAHEK LKVGSVDVLH CYYAHGEDNE NFQRRCYWML 120 EQDLMHIVFV HYLEVQGNKS NVGDRESNEV TSNPGKHSSL TFSFPGNRTK APSGITDSTS 180 PTSTLTLSCE DADSGYDAED SHQASSRAHL YYELPQMGNG PRMEKMDSGL SYSYFLSPSS 240 GCPEVRSSIP GDYVSHAGHI PNDNQDLMIE CQKALGLASW EEVLEHCSGE NDNVPPHAKL 300 ESNVQKENIF DGELLSREAS EENSGSSLPV QFNWQIPLAD NSSHFLKSIM DLSRDLEPAY 360 DLGDGLFEQR THDACLLGAP EPFCAFLDQQ NELPVQNNLQ MQQRDVESHS QTKSNSESEI 420 HGEGTINFSF SVKQKLLNGE GNLEKVDSFS RWMSKELEEV DNLHVQSSGI EWSTEECGNV 480 VDDSSLSPSL SQDQLFSIID FSPKWTYTDP EIEVVVTGMF LKSHQEVAKC KWSCMFAEVE 540 VPAEVLADGV LCCRIPPHAV GRVPFYITCS NRLACSEVRE FDYIVGSVKD ADISDIYGSS 600 TSESFLHLRL ERILSMRSSP QNHLSEGLCE KQKLISKIIQ LKEEEESYQM VEANPEKNLS 660 QHVEKYQILQ KIMKEKLYSW LLRKVCEDGK GPCILDDEGQ GVLHLAASLG YDWAIKPTVT 720 AGVSINFRDL SGWTALHWAA YCGREKTVAV LLSLGAAPGL LTDPSPEFPL SRTPSDLASS 780 NGHKGISGFL AESSLTSLLL SLKMNDSADD GALEDSIAKA VQTVSEKTAT PANDNDESDV 840 LSLKDSLTAI CNATQAADRI HQIFRMQSFQ RKQLTEFNNE LGISYEHALS LVAAKSLRPV 900 QGDGLAHSAA IQIQKKFRGW KKRKEFLLIR QRIVKIQAHV RGHQARKKYR PIIWSVGILE 960 KVILRWRRKG SGLRGFRRDA LGMNPNPQHM PLKEDDYDFL KDGRKQTEER LQKALGRVKS 1020 MVQYPEARAQ YRRLLTVVEG SRETKQESNM VPNGLEDIAD GDLDLIDIDS LLDDDTFMSV 1080 AFE* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.2613 | 1e-75 | vegetative meristem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, old leaves, petals, sepals, top of carpels, stigmas, stamen filaments, anthers and siliques, but not in pollen. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
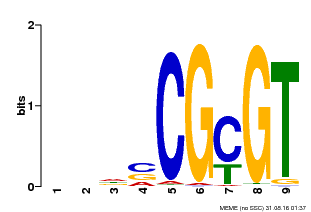
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015388622.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A2H5QEH7 | 0.0 | A0A2H5QEH7_CITUN; Uncharacterized protein | ||||
| STRING | XP_006487645.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g001406m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



