 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g001436m | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1079aa MW: 121191 Da PI: 5.4939 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188 | 9.1e-59 | 21 | 136 | 4 | 118 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktvrE+hekLKvg+v+vl+cyYah+e+n++f
orange1.1g001436m 21 EaQHRWLRPAEICEILCNYQKFHIASEPPSRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVREAHEKLKVGSVDVLHCYYAHGEDNENF 112
449***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrrcyw+Le++l +iv+vhylev+
orange1.1g001436m 113 QRRCYWMLEQDLMHIVFVHYLEVQ 136
*********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.221 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.0E-85 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.4E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.8E-5 | 478 | 579 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.01E-14 | 494 | 579 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 1.2E-16 | 676 | 789 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.37E-16 | 682 | 788 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.82E-13 | 683 | 786 | No hit | No description |
| PROSITE profile | PS50297 | 16.255 | 694 | 788 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0042 | 727 | 756 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.698 | 727 | 759 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.45E-8 | 901 | 952 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.099 | 901 | 923 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.151 | 902 | 931 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 9.1E-4 | 903 | 922 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.002 | 924 | 946 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.725 | 925 | 949 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.4E-4 | 927 | 946 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1079 aa Download sequence Send to blast |
MADRGSYALA PRLDMQQLQM EAQHRWLRPA EICEILCNYQ KFHIASEPPS RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVREAHEK LKVGSVDVLH CYYAHGEDNE NFQRRCYWML 120 EQDLMHIVFV HYLEVQGNKS NVGDRESNEV TSNPGKHSSL TFSFPGNRTK APSGITDSTS 180 PTSTLTLSCE DADSGYDAED SHQASSRAHL YYELPQMGNG PRMEKMDSGL SYSYFLSPSS 240 VRSSIPGDYV SHAGHIPNDN QDLMIECQKA LGLASWEEVL EHCSGENDNV PPHAKLESNV 300 QKENIFDGEL LSREASEENS GSSLPVQFNW QIPLADNSSH FLKSIMDLSR DLEPAYDLGD 360 GLFEQRTHDA CLLGAPEPFC AFLDQQNELP VQNNLQMQQR DVESHSQTKS NSESEIHGEG 420 TINFSFSVKQ KLLNGEGNLE KVDSFSRWMS KELEEVDNLH VQSSGIEWST EECGNVVDDS 480 SLSPSLSQDQ LFSIIDFSPK WTYTDPEIEV VVTGMFLKSH QEVAKCKWSC MFAEVEVPAE 540 VLADGVLCCR IPPHAVGRVP FYITCSNRLA CSEVREFDYI VGSVKDADIS DIYGSSTSES 600 FLHLRLERIL SMRSSPQNHL SEGLCEKQKL ISKIIQLKEE EESYQMVEAN PEKNLSQHVE 660 KYQILQKIMK EKLYSWLLRK VCEDGKGPCI LDDEGQGVLH LAASLGYDWA IKPTVTAGVS 720 INFRDLSGWT ALHWAAYCGR EKTVAVLLSL GAAPGLLTDP SPEFPLSRTP SDLASSNGHK 780 GISGFLAESS LTSLLLSLKM NDSADDGALE DSIAKAVQTV SEKTATPAND NDESDVLSLK 840 DSLTAICNAT QAADRIHQIF RMQSFQRKQL TEFNNELGIS YEHALSLVAA KSLRPVQGDG 900 LAHSAAIQIQ KKFRGWKKRK EFLLIRQRIV KIQAHVRGHQ ARKKYRPIIW SVGILEKVIL 960 RWRRKGSGLR GFRRDALGMN PNPQHMPLKE DDYDFLKDGR KQTEERLQKA LGRVKSMVQY 1020 PEARAQYRRL LTVVEGSRET KESNMVPNGL EDIADGDLDL IDIDSLLDDD TFMSVAFE* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.2613 | 4e-82 | vegetative meristem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, old leaves, petals, sepals, top of carpels, stigmas, stamen filaments, anthers and siliques, but not in pollen. {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
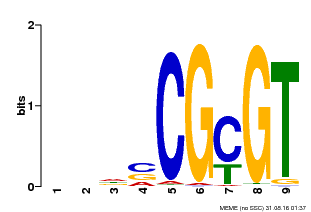
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006487646.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X3 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A2H5QED5 | 0.0 | A0A2H5QED5_CITUN; Uncharacterized protein | ||||
| STRING | XP_006487645.1 | 0.0 | (Citrus sinensis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g001436m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



