 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g001453m | ||||||||
| Common Name | CISIN_1g001317mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1076aa MW: 118777 Da PI: 8.227 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130 | 8.9e-41 | 145 | 222 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCe hsk++++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
orange1.1g001453m 145 MCQVDNCKEDLSNAKDYHRRHKVCELHSKSTKALVGKQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 222
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.8E-34 | 141 | 207 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.855 | 143 | 220 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-37 | 144 | 225 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.6E-29 | 146 | 219 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1076 aa Download sequence Send to blast |
MTMTMTMAKK RHLSYQAQSQ NHYGGEQQNW NPKLWDWDSV GFVGKPVVDS DPEVLRLGGA 60 TASESPNKTT DNINYNYNYN NQKKGNTTTT SAVTVGNVED DGRLDLNLGG GLTAVDVEQP 120 EPPVVTSKPN KRVRSGSPGT APYPMCQVDN CKEDLSNAKD YHRRHKVCEL HSKSTKALVG 180 KQMQRFCQQC SRFHPLSEFD EGKRSCRRRL AGHNRRRRKT QPEDITSRML IHGHGNQSNN 240 PTANVDIVNL LTALARAQGK TEDRSISCSS VPDREQLLMI LSKINSLPLP ADLAAKLHNF 300 GSLNRKTPVH TSTDVQNRLN ENTSSPSTMD LLAVLSSTLT APSPDTLAAH SQRSSHSSDS 360 EKTKSTCPEQ ATPNFLKRTT MDFPSVGGER SSTSYQSPVE DSDGQNQETR VNLPLQLFSS 420 SPEDDSPPKL SSSRKYFSSD SSNPIEERSP SSSPVVQTFF PMQSTSETVK SEKLSIGREV 480 NANVEGNRSR GSIMPLELFR GSNKAADNCS FQSFPYQAGY TSSSGSDHSP SSLNSDAQDC 540 TGRIIFKLFD KDPSQFPGTL RKEIYNWLSN SPSEMESYIR PGCVILSLYV SMPYATWEQL 600 EGNLLQRINS LVQDSDSDFW RNARFLVHTG KQLASHKDGN IRVCKSWRTW SSPELISVSP 660 LAVVGGQELS FKLRGRNLTN LGTKIHCTFM GGYASQEVTS STCQGSIYDE IILAGLKIQD 720 TSPSVLGRFF IEVENGFKGN SFPVIIADAT ICKELSLLES EFGAEAKVCD VISEHQAHEY 780 GRPRSREEVL HFLNELGWLF QRKRASSIVK GSDYSLSRFK FLLVFSVDRG CCALVKAILD 840 ILVEGNLSMD GLSRESLEML WEIQLLNRAV KMKCRRMVDL LIHYSLTSSN DTPQKYIFPP 900 NLAGPGGITP LHLAACTSDS DDIIDALTND PQEIGPSSWN SILDASGHSP YSYALMKNNH 960 AYNKLVARKL ADRRNGQVTI PVGVEIEQSG LAKEQVHGLS SQFKQRGKSC TKCAVAAAKL 1020 NKRVRGSQGL LNRPYIHSML AIAAVCVCVC LFLRGSPDIG LVAPFKWENL DFGPK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-31 | 136 | 219 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.10084 | 1e-161 | callus | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in vasculature of aerial parts. Expressed in cotyledons and leaf petioles. {ECO:0000269|PubMed:15703061}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
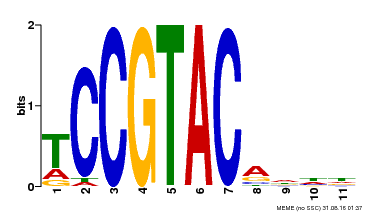
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006435483.1 | 0.0 | squamosa promoter-binding-like protein 14 isoform X1 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A067H2Z6 | 0.0 | A0A067H2Z6_CITSI; Uncharacterized protein | ||||
| TrEMBL | V4SLU4 | 0.0 | V4SLU4_9ROSI; Squamosa promoter-binding protein 12 | ||||
| STRING | XP_006435483.1 | 0.0 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g001453m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



