 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g002122m | ||||||||
| Common Name | CISIN_1g001971mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 965aa MW: 107018 Da PI: 6.692 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132.4 | 1.5e-41 | 149 | 226 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adls+ak+yhrrhkvCe+hska+ +lv +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
orange1.1g002122m 149 VCQVEDCGADLSNAKDYHRRHKVCEMHSKASRALVGNVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 226
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 8.9E-34 | 142 | 211 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.385 | 147 | 224 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.41E-39 | 148 | 229 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.3E-30 | 150 | 223 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 5.2E-5 | 304 | 333 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 7.93E-6 | 737 | 849 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 5.2E-5 | 738 | 848 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.06E-6 | 738 | 849 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 965 aa Download sequence Send to blast |
METRFRGEAH HFYGMNSMDL RAVGKKTLEW DLNDWKWDGD LFIASKLNPA PNENIGRQFF 60 PLAVGNSSNS SSSCSDEVNL GIENGKREVE KKRRAVVVED HNSYEVAAGG LSLKLGGNGH 120 PLSEREMGNW AGSSGKKTKF GGGSSSRAVC QVEDCGADLS NAKDYHRRHK VCEMHSKASR 180 ALVGNVMQRF CQQCSRFHVL QEFDEGKRSC RRRLAGHNKR RRKTNPDAVA NGSSPNNDQT 240 SGYLLISLLR ILSNMHSSRS DQRTDQDLLS HLLRGLASPA GENGGRGISG LLQEHQDMLN 300 ERTSAGNSEV VQAFLANGQG CPTPFRQQLN ATVSEMPQQV SLPHDARGAE DQDEDVERSP 360 VPANLGTSSI DCPSWVRQDS QQSSPPQTSG NSDSASAQSP SSSSDAQSRT DRIVFKLFGK 420 EPNDFPLVLR AQILDWLSHS PSDMESYIRP GCVILTIYLR QAEAAWEELC CDLTFSLSRL 480 LDLSNDSFWT SGWVYARVQH QIAFIYNGQV VLDTSLPPRS NNYSKILSVK PIAVPASERA 540 QFFVKGINLG RSATRLLCAV EGKYMVQEAT HELLDDVDGF KELDELQCVN FSCSIPAVTG 600 RGFIEIEDHG FSSTFFPFIV AEEDVCSEIR MLESALEFNR TDADVERFGK IDTKNQAMDF 660 IHEIGWLFHR SQSKSRLGHL DPNTDLFPLR RFKWLIEFSM DHEWCAVVKK LLHILLDGTV 720 SLGEHPSLDL ALTELGLLHR AVRKNSRPLV DLLLRFVPLE VSDRLGSENK ALVDGVHKGF 780 LFRPDVIGPA GLTPIHIAAG KDGSEDVLDA LTDDPGMVGI EAWKNARDSS GSTPEDYARL 840 RGHYSYIHLV QKKINKRPNG GHVVVDICGV VPDSNIYQKQ NNESTASFEI GQTPVRPTQH 900 NCKLCHQKLG YATASRSLVY KPAMLSMVAI AAVCVCVALL FKSCPEVLYV FRPFRWEMLD 960 YGTS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-31 | 143 | 223 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.11650 | 0.0 | callus | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed constitutively during plant development. {ECO:0000269|PubMed:10524240}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
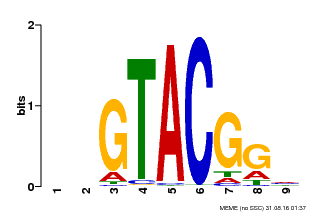
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006444595.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A067H7A4 | 0.0 | A0A067H7A4_CITSI; Uncharacterized protein | ||||
| STRING | XP_006444595.1 | 0.0 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g002122m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



