 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g002516m | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 914aa MW: 101174 Da PI: 5.2285 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 77.3 | 1.6e-24 | 114 | 215 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkld 85
f+k+lt sd++++g +++p++ ae+ ++++ + ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv +++L++gD+v+F
orange1.1g002516m 114 FCKTLTASDTSTHGGFSVPRRAAEKLfppldytMQPP-T-QELVVRDLHDNTWTFRHIYRGQPKRHLLTTGWSLFVGSKRLRAGDSVLFI-- 201
99*********************999*****954444.4.38************************************************.. PP
SSSEE..EEEEE-S CS
B3 86 grsefelvvkvfrk 99
++++ +l+v+v+r+
orange1.1g002516m 202 RDEKSQLMVGVRRA 215
4577778*****97 PP
| |||||||
| 2 | Auxin_resp | 115.5 | 4.5e-38 | 240 | 323 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaas++s F+++YnPra++s+Fv++++k++k+++ +++svGmRf m+fete+s +rr++Gt+vg+sdldp+rWp+SkWr+L+
orange1.1g002516m 240 AAHAASNRSQFTIFYNPRACPSDFVIPLAKYRKSVYgTQMSVGMRFGMMFETEESGKRRYMGTIVGISDLDPLRWPGSKWRNLQ 323
79**********************************9*********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 7.32E-47 | 101 | 243 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 9.6E-42 | 107 | 228 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 4.02E-23 | 113 | 214 | No hit | No description |
| SMART | SM01019 | 3.0E-25 | 114 | 216 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.1E-22 | 114 | 215 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.211 | 114 | 216 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 4.1E-33 | 240 | 323 | IPR010525 | Auxin response factor |
| SuperFamily | SSF54277 | 4.91E-10 | 798 | 878 | No hit | No description |
| Pfam | PF02309 | 3.6E-10 | 799 | 884 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.619 | 799 | 883 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 914 aa Download sequence Send to blast |
MGSVEEKIKA GGLVIRAQTT LLEEMKLLKE MQDQSGARKA INSELWHACA GPLVFLPQVG 60 SLVYYFPQGH SEQADKDTDE IYAQMSLQPV NSEKDVFPIP DFGLKPSKHP SEFFCKTLTA 120 SDTSTHGGFS VPRRAAEKLF PPLDYTMQPP TQELVVRDLH DNTWTFRHIY RGQPKRHLLT 180 TGWSLFVGSK RLRAGDSVLF IRDEKSQLMV GVRRANRQQT ALPSSVLSAD SMHIGVLAAA 240 AHAASNRSQF TIFYNPRACP SDFVIPLAKY RKSVYGTQMS VGMRFGMMFE TEESGKRRYM 300 GTIVGISDLD PLRWPGSKWR NLQVEWDEPG CSDKQKRVSP WEIETPESLF IFPSLTSGLK 360 RPFHSGILAT ETEWGSLIKR PLACPEIAPG VMPYSSISNL CSEQLIKMML KPQLVNNPGS 420 FAASSLQETS GAKGAHLEEV KTLQSTINQK PRLVPSEMNR IDNQNCSQIC LNQADTVNSS 480 LSRINIPEKP HPPSKCEKQA PPGMNTDHLK SEPRQSIEQS SNLTSAADCS MEKPSGPLNP 540 QNLVNQHAFH NQNEGLLQLQ SSWPMQSQLE SVFQAQQINV PQSDSTAHSG SLPILDTDEW 600 MSHTSCNSLA GTYNRSPGPL PMFGLQEPST MLPEVINPSL SFPGQEMWDH QLNNLRFLSP 660 VDPLTSFTQQ DHCSLNSSGL RDLSDESNNQ SGIYSCLNID VSNGGSTMID HSVSSAILDE 720 FCTLKDANFQ NPPDCLMNTF SSSQDVQSQI TSASLADSQA FSRQDFPDNS GGTSSSNVDF 780 DESSLLQNTS WQPVVPPMRT YTKVQKTGSV GRSIDVTNFK NYDELCSAIE RMFGLEGLLN 840 DPRGTEWKLV YVDYENDVLL VGDDPWEEFV GCVRCIRILS PQEVEQMSEE GMKLLNSAAM 900 QGIDCTKPEG GRA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 348 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.11086 | 0.0 | flower | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In early embryo and during organ development. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant with a lower expression in leaves. Detected in embryo axis, provascular tissues, procambium and some differentiated vascular regions of mature organs. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
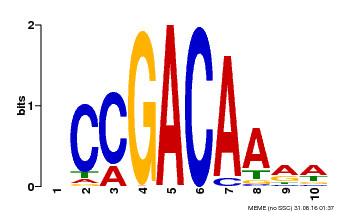
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024039097.1 | 0.0 | auxin response factor 5 isoform X1 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | V4TFD4 | 0.0 | V4TFD4_9ROSI; Auxin response factor | ||||
| STRING | XP_006435146.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5808 | 27 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g002516m |



