 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g006602m | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 640aa MW: 71296.1 Da PI: 8.3164 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 41.8 | 2.9e-13 | 22 | 51 | 49 | 78 |
EEETTT--SS--S-STTTT-------S--- CS
SBP 49 fhelsefDeekrsCrrrLakhnerrrkkqa 78
fh ls+fDe+krsCrr+L++hn+rrr+k+
orange1.1g006602m 22 FHLLSDFDEGKRSCRRKLERHNNRRRRKSV 51
9*************************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 12.255 | 1 | 49 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.71E-10 | 22 | 52 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 9.3E-7 | 22 | 49 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 640 aa Download sequence Send to blast |
MRKRFYRFAR WREQEILSTV WFHLLSDFDE GKRSCRRKLE RHNNRRRRKS VDSKGAVDSE 60 PPGASRCEDI ICDDDSGKDS LCLSSQITDQ EAFLESEDGL VSALNSAPNT QNVKSDSGIS 120 AVASGEIRTD RGKDDSKASL SPSNCDNKSS YSSLCPTGRI SFKLYDWNPA EFPRRLRHQI 180 FHWLASMPVE LEGYIRPGCT ILTVFIAMPK IMWAKLYEDP IRYVHNFVVE PGTASMLSGR 240 GSMFVHLNNM IFHVKGGTSV VKVDVKVQAP KLHYVQPSCF EAGKPLEFVA CGSNLIQPKL 300 RYVTYLPHDY CIVSPLGGSE GESLALEHQF YKINVPHIEA NLFGPAFIEV ENESGLSNFI 360 PVLIGDKGTC SEINIIQQRF EASFFSKRSQ FMASGLLSDL CEVSALRQKA LTELLVDIAW 420 LLKAPASESF RQTISCSEVQ RFNQLLSFLI YNESTTILEK MLQNMKILMN NIESNIAVNG 480 ISDSDMGLLL KYMDYARGIL CQKVKKDKGP MQHSGNIVPK MISSSQSCLQ ANSLVPSTKQ 540 DLVVRSNDKI GAVMGSATVD RCEVVPLLNR EVVMNVNLIK EWPRKSCSPI FSGRVLSSLP 600 TVTVIAMAAV CFGVCLVVLH PQKVGHFATS IRRSLFGNL* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 35 | 47 | RRKLERHNNRRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
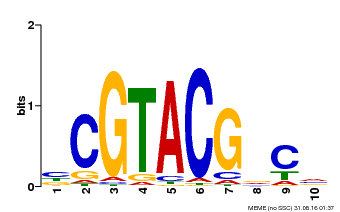
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006419725.1 | 0.0 | squamosa promoter-binding-like protein 7 isoform X2 | ||||
| TrEMBL | V4S2K0 | 0.0 | V4S2K0_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006419724.1 | 0.0 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.1 | 0.0 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g006602m |



