 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g007662m | ||||||||
| Common Name | CISIN_1g007662mg, LOC102616379 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 595aa MW: 67093 Da PI: 5.4294 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 438.6 | 5.7e-134 | 37 | 379 | 1 | 320 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLr 92
eel++rmwkd+++lkr+ke++k +++ a++++k +++++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+g+sd++r
orange1.1g007662m 37 EELERRMWKDRIKLKRIKEKEKLA--AQQ-AAEKQKPKQTTDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGSSDNIR 125
8**************999999963..355.599*********************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHH CS
EIN3 93 aWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgel 184
aWWkekv+fd+ngpaai+ky+a++l++s++++++ + ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekgv+pPWWP+G+e+ww++l
orange1.1g007662m 126 AWWKEKVKFDKNGPAAIAKYEAECLAMSEADNNNR-NGNSQSILQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGVPPPWWPSGNEDWWEKL 216
****************************9998865.8889999************************************************* PP
T--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.........XX CS
EIN3 185 glskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.........sl 267
gl+k q ppykkphdlkk+wkv+vLtavikhmsp+i++ir+++rqsk+lqdkm+akes+++l vl++ee+++++ s+++ +
orange1.1g007662m 217 GLPKGQI-PPYKKPHDLKKMWKVGVLTAVIKHMSPDIAKIRRHVRQSKCLQDKMTAKESAIWLGVLSREESLIRQPSSDNGtsgitevpsGV 307
******9.9**********************************************************************4445677777544 PP
XXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX......................XXXXXXXXXXXXXXXXXX CS
EIN3 268 rkqspkvtlsceqkedvegkkeskikhvqavktta......................gfpvvrkrkkkpsesakv 320
+ ++++ +s+++++dv+g ++ + ++++++t+ + +kr+ + ++++ +
orange1.1g007662m 308 HGNKKQPAISSDSDYDVDGIDDVVGSVSSKDNRTNqpvdvepvishhnatihsfqdkE--RPEKRR-RAKRPRVT 379
558889999*********8888888999999998899999999999977666554322..222222.22222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 6.0E-127 | 37 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 5.5E-72 | 159 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 8.24E-62 | 166 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 595 aa Download sequence Send to blast |
MGELEEIGAD ICSDIEVDER CGNLVEKDVS DEEIEPEELE RRMWKDRIKL KRIKEKEKLA 60 AQQAAEKQKP KQTTDQARRK KMSRAQDGIL KYMLKLMEVC KARGFVYGII PEKGKPVSGS 120 SDNIRAWWKE KVKFDKNGPA AIAKYEAECL AMSEADNNNR NGNSQSILQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKGV PPPWWPSGNE DWWEKLGLPK GQIPPYKKPH DLKKMWKVGV 240 LTAVIKHMSP DIAKIRRHVR QSKCLQDKMT AKESAIWLGV LSREESLIRQ PSSDNGTSGI 300 TEVPSGVHGN KKQPAISSDS DYDVDGIDDV VGSVSSKDNR TNQPVDVEPV ISHHNATIHS 360 FQDKERPEKR RRAKRPRVTS SSADQQPGES VNEHQDDEPR KTLLDINHSD VLLIDYNMQG 420 NQEQNNAVTA LRPVEGGFVG HSQISASEFN NFSAIPSVDI MATQSMYVDG RPTVYPMMQN 480 TELQHDDTCN FYNPSFEFGH SNDGQHGQIE TNVPQIRPEV GGVHVPAFHR NENEITTNGG 540 EMPHYINNTF HSEQERNFDH QFGSPLDVMP WMEGSFDDII VDDFDDDLLK YFGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-75 | 165 | 296 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 367 | 375 | EKRRRAKRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
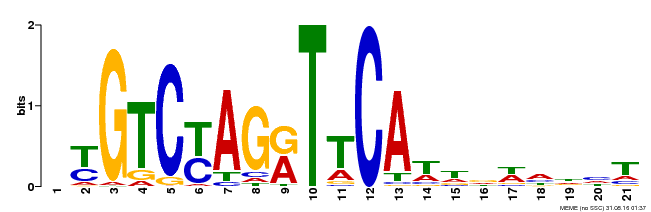
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006494725.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A067FBX3 | 0.0 | A0A067FBX3_CITSI; Uncharacterized protein | ||||
| TrEMBL | A0A2H5PBY8 | 0.0 | A0A2H5PBY8_CITUN; Uncharacterized protein | ||||
| STRING | XP_006494725.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-172 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g007662m |
| Entrez Gene | 102616379 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



