 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g010039m | ||||||||
| Common Name | CISIN_1g010039mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 520aa MW: 57081.8 Da PI: 6.2892 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 45.6 | 1.8e-14 | 251 | 310 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + g ++ ++++lg ++ +++Aa+a++ a++k++g
orange1.1g010039m 251 SIYRGVTRHRWTGRYEAHLWDnSCRrEGqaRKgRQVYLGGYDKEDKAARAYDLAALKYWG 310
57*******************666664478446**********99*************98 PP
| |||||||
| 2 | AP2 | 39.5 | 1.4e-12 | 342 | 387 | 7 | 55 |
AP2 7 rwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
r+++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
orange1.1g010039m 342 RRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 387
89**********988532...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 5.77E-22 | 251 | 320 | No hit | No description |
| SuperFamily | SSF54171 | 3.6E-17 | 251 | 320 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.1E-11 | 251 | 310 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.7E-16 | 252 | 319 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.137 | 252 | 318 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.6E-27 | 252 | 324 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-6 | 253 | 264 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.621 | 328 | 395 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.6E-7 | 341 | 387 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.7E-16 | 343 | 395 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-15 | 343 | 396 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 2.9E-24 | 343 | 401 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-6 | 377 | 397 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010080 | Biological Process | regulation of floral meristem growth | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0035265 | Biological Process | organ growth | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 520 aa Download sequence Send to blast |
MAPASNWLSF SLSPMEMWRS SSSSDSQFLP YDASQSAASP HYFLDNFYAN AAGWTNPKSQ 60 VFFTEEENIQ AKELEVAAAQ TMAADSSILP SLTQTHPVPK LEDFFADSSS LVHSQTETQD 120 SSLTHIYDHH HHGSAYFNDH QQDLKAIAGF QAFSTNSGSE VDDSTSMTRT QLPAAEFVGH 180 SIESTGNELG FSNSCPTTGC LSLGVNNNAT NNANSNNNVN KNQGCNEKKA IVSADSDCTK 240 KVADTFGQRT SIYRGVTRHR WTGRYEAHLW DNSCRREGQA RKGRQVYLGG YDKEDKAARA 300 YDLAALKYWG PTATTNFPVS GYTKELEEMK HVTKQEFIAS LRRHHQQGRW QARIGRVAGN 360 KDLYLGTFAT EEEAAEAYDI AAIKFRGANA VTNFEMNRYD VEAIGKSSLP IGGAAKRLKL 420 SLESEQKLAL TQDQQPQCSS SSNNINFAAL QPVGPASIPC GIPFDAATAA ALYHQNLYHH 480 FQTTNFNPAD SPGSTSSMAP PVALMQQPAE FFLWPHQSY* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Detected in inflorescence and youg floral mersitems, and in stem procambial cells. In floral mersitems, mostly expressed in the central dome. Disappears progressively from sepal primordia, but accumulates in second, third and fourth whorl organ primordia. Later, confined to occasional patches in stamens and in petal before disparearing progressively from flowers. {ECO:0000269|PubMed:15988559}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, seedlings, hypocotyl, inflorescence, siliques, and pistils. Also detected at low levels in leaves. {ECO:0000269|PubMed:15988559, ECO:0000269|PubMed:16307362}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00504 | DAP | Transfer from AT5G10510 | Download |
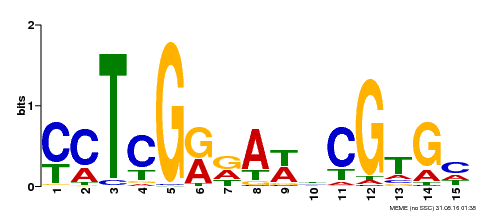
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024036659.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL6 isoform X3 | ||||
| Swissprot | Q52QU2 | 1e-178 | AIL6_ARATH; AP2-like ethylene-responsive transcription factor AIL6 | ||||
| TrEMBL | A0A067F341 | 0.0 | A0A067F341_CITSI; Uncharacterized protein | ||||
| STRING | XP_006466326.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G10510.2 | 1e-176 | AINTEGUMENTA-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g010039m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



