 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g011009m | ||||||||
| Common Name | CISIN_1g010454mg, LOC102606994 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 496aa MW: 53509.7 Da PI: 6.7944 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.4 | 6.3e-17 | 171 | 220 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
orange1.1g011009m 171 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 220
78*******************......55************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 5.7E-9 | 171 | 220 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.31E-15 | 171 | 229 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 6.5E-17 | 172 | 228 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.794 | 172 | 228 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.5E-31 | 172 | 234 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.72E-11 | 172 | 227 | No hit | No description |
| SuperFamily | SSF54171 | 2.16E-6 | 263 | 307 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 4.89E-13 | 263 | 308 | No hit | No description |
| SMART | SM00380 | 5.7E-13 | 264 | 312 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 496 aa Download sequence Send to blast |
MLDLNLNVMS IESTQQNSDE ERVGSVVLVE KSPEGSSGTS NSSVVNADAS SNVGDDESCS 60 TRAVAGDNSV FTFNFDILKV GGDSGNVRNE NVEQNNAASR KEFAFFPASG GGENGGGQSS 120 GSWIDLSFDK QQQQYQNQQP QQQEGGGEVR VTQQSSPVVK KSRRGPRSRS SQYRGVTFYR 180 RTGRWESHIW DCGKQVYLGG FDTAHAAARA YDRAAIKFRG VDADINFNLA DYEDDMKQMK 240 NLTKEEFVHI LRRQSTGFSR GSSKYRGVTL HKCGRWEARM GQFLGKKAYD KAAIKCNGRE 300 AVTNFEPSTY EGEMITEGSN EGGDHNLDLN LGISSSFGSG PKANEGHSQF LSGPYGAHGG 360 RTSRTENSTT AAVGDSASKG LMRTSEHPIF WNGAYPGLLT TEEKATEKRI ELGPPQGLPN 420 WAWQMHGQVS GTPASLLSSA ASSGFPFSAA PPSAAVLSSK PLNPSDVNLC FTSTNTTAPN 480 TSQFYYQMKL PQAPP* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.17126 | 0.0 | callus| flower| fruit | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
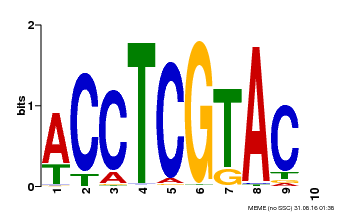
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006488204.1 | 0.0 | ethylene-responsive transcription factor RAP2-7 isoform X2 | ||||
| Swissprot | Q9SK03 | 1e-123 | RAP27_ARATH; Ethylene-responsive transcription factor RAP2-7 | ||||
| TrEMBL | A0A067G3U2 | 0.0 | A0A067G3U2_CITSI; Uncharacterized protein | ||||
| STRING | XP_006424678.1 | 0.0 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.1 | 1e-113 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g011009m |
| Entrez Gene | 102606994 |



