 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g017035m | ||||||||
| Common Name | CISIN_1g017035mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 379aa MW: 42006.4 Da PI: 9.2546 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 45.8 | 1.5e-14 | 69 | 117 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f +eeAa+a++ a+++++g
orange1.1g017035m 69 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEEEAARAYDIAAQRFRG 117
89****9888.8*********3.....5*************************98 PP
| |||||||
| 2 | B3 | 101.6 | 4.4e-32 | 207 | 306 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+ tpsdv+k++rlv+pk++ae+h +g++ ++ l +ed +g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++
orange1.1g017035m 207 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqSGSTSKGLLLNFEDVTGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDIVSFHRSTGG 298
89****************************77888999*************************************************87667 PP
EE..EEEE CS
B3 89 efelvvkv 96
+++l++ +
orange1.1g017035m 299 DRQLYIDW 306
77777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 2.2E-9 | 69 | 117 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-18 | 69 | 125 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.74E-27 | 69 | 124 | No hit | No description |
| PROSITE profile | PS51032 | 20.653 | 70 | 125 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.0E-21 | 70 | 126 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.5E-30 | 70 | 131 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.8E-5 | 71 | 82 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.8E-5 | 91 | 107 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 1.5E-40 | 202 | 309 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.88E-30 | 206 | 307 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 9.41E-29 | 206 | 304 | No hit | No description |
| PROSITE profile | PS50863 | 14.48 | 207 | 310 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.9E-25 | 207 | 310 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.5E-29 | 207 | 307 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 379 aa Download sequence Send to blast |
MKREEMDGSC MDESTTTDSA STSPPITFPA SIPPTKSPES LCRVGSGASS VILDSEAGVE 60 AESRKLPSSK YKGVVPQPNG RWGAQIYEKH QRVWLGTFNE EEEAARAYDI AAQRFRGRDA 120 VTNFKQISCA GTSSDDGDIE MAFLSSHSKS EIVDMLRKHT YNDELEQSKR NYGLDANGKR 180 VIKHGEGDGA ATAFGSDRVL KARDQLFEKA VTPSDVGKLN RLVIPKQHAE KHFPLQSGST 240 SKGLLLNFED VTGKVWRFRY SYWNSSQSYV LTKGWSRFVK EKNLKAGDIV SFHRSTGGDR 300 QLYIDWKART GPIENPVEPV QMMRLFGVNI FKIPGNGLVG VDKIGCNINN NNNNGKRLRE 360 MELLSLECSK KQRMIGAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 6e-59 | 204 | 311 | 11 | 119 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Csi.562 | 0.0 | flower| fruit | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences (Probable). Functionally redundant with TEM1. {ECO:0000250, ECO:0000269|PubMed:18718758, ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
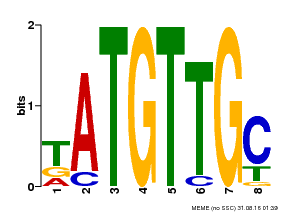
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006437734.1 | 0.0 | AP2/ERF and B3 domain-containing transcription repressor RAV2 | ||||
| Swissprot | P82280 | 1e-156 | RAV2_ARATH; AP2/ERF and B3 domain-containing transcription repressor RAV2 | ||||
| TrEMBL | A0A067FRT4 | 0.0 | A0A067FRT4_CITSI; Uncharacterized protein | ||||
| STRING | XP_006437734.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM848 | 27 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-158 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g017035m |



