 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g017566m | ||||||||
| Common Name | CISIN_1g017566mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 370aa MW: 40219.1 Da PI: 5.3339 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.6 | 1.9e-15 | 59 | 104 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++e d l ++v ++G ++W++Iar + gR++k+c++rw +
orange1.1g017566m 59 KGPWSPEQDAVLSELVSKFGARNWSLIARGIA-GRSGKSCRLRWCNQ 104
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 56.2 | 7.6e-18 | 111 | 155 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r+++T eEd+++v a++ +G++ W+ I+r ++ gRt++ +k++w++
orange1.1g017566m 111 RKPFTDEEDQIIVAAHAVHGNK-WAVISRLLP-GRTDNAIKNHWNST 155
789*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 19.009 | 54 | 105 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.49E-29 | 56 | 152 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.6E-13 | 58 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-14 | 59 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-24 | 60 | 112 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.85E-12 | 61 | 103 | No hit | No description |
| PROSITE profile | PS51294 | 25.217 | 106 | 160 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.0E-16 | 110 | 158 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.1E-15 | 111 | 155 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-23 | 113 | 159 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.19E-12 | 113 | 156 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 370 aa Download sequence Send to blast |
MKMEPETEIE KWKATVIEVE GGDAVEADAD GGADRGRSGD DSVVAVGEGS KRSGRSRVKG 60 PWSPEQDAVL SELVSKFGAR NWSLIARGIA GRSGKSCRLR WCNQLDPAVK RKPFTDEEDQ 120 IIVAAHAVHG NKWAVISRLL PGRTDNAIKN HWNSTLRRRG VELRRTMKLG SVNMMEDMNV 180 DKTKASSEET LSCGDVNSFK FLEEKDTSSL ENLNNHCGDK STTEVLSSQE TEEPPTLFRP 240 LARVSAFSVY NTLDDPETAS IHPRQTPSQG PLVQVTMPDA GTCKLLEGVY GERMVPHWCG 300 YGCCGIQHGG NCQSSLLGPE FFDFSGPPAF PSYELAAIAT DISNVAWLKS GLENSSVTVM 360 GDEAGRIKN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 4e-39 | 58 | 159 | 6 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00337 | DAP | Transfer from AT3G09230 | Download |
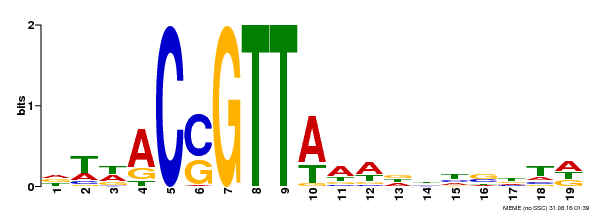
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid. {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006480929.1 | 0.0 | transcription factor MYB1 | ||||
| Swissprot | Q42575 | 1e-108 | MYB1_ARATH; Transcription factor MYB1 | ||||
| TrEMBL | A0A067ESC3 | 0.0 | A0A067ESC3_CITSI; Uncharacterized protein | ||||
| STRING | XP_006480929.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8492 | 26 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09230.1 | 1e-111 | myb domain protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g017566m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



