 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g024702m | ||||||||
| Common Name | CISIN_1g021851mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 265aa MW: 29023.6 Da PI: 8.3217 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 22.3 | 3.1e-07 | 2 | 28 | 19 | 47 |
TTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 19 lGggtWktIartmgkgRtlkqcksrwqky 47
+G g W+ I +++g ++t+ q++s+ qk+
orange1.1g024702m 2 YGRG-WRQIEEHVG-TKTAVQIRSHAQKF 28
8999.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.697 | 1 | 33 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.44E-5 | 2 | 32 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 0.00382 | 2 | 29 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 265 aa Download sequence Send to blast |
MYGRGWRQIE EHVGTKTAVQ IRSHAQKFFS KVVRESNGSS ESSIMPIEIP PPRPKRKPVH 60 PYPRKSVDSL KATSVSNQQE NFTSSNALVS DKDRQSPTSV VSAFNSDTLG CAASDQQNGC 120 SSPTSCTTEM HSVNLLPIEK ENEYVTSISF PKEEKISTLP AHLSASSNVE ELASVSKDSV 180 YPKGDAAAAP SCTSIKLFGR TVLVSDSWKP YSLGADSYKS PISKSSQENL DVDKKILSSQ 240 PHQSIWIHIY YLGWFLAIVI HPHI* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
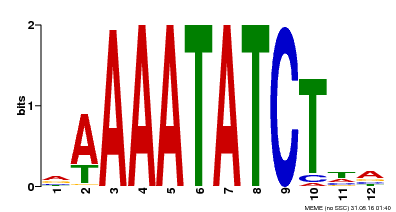
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006441350.1 | 1e-157 | protein REVEILLE 7 isoform X1 | ||||
| TrEMBL | A0A067DR78 | 0.0 | A0A067DR78_CITSI; Uncharacterized protein | ||||
| TrEMBL | A0A067DYI8 | 0.0 | A0A067DYI8_CITSI; Uncharacterized protein | ||||
| STRING | XP_006441350.1 | 1e-156 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 2e-27 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g024702m |



