 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g026888m | ||||||||
| Common Name | CISIN_1g020302mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 232aa MW: 26060.4 Da PI: 7.5719 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 74.5 | 1.8e-23 | 181 | 228 | 1 | 48 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSS CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsr 48
+Cq+egC+adl++ak+yhrrhkvCe+hska++v+++gl+qrfCqqCsr
orange1.1g026888m 181 RCQAEGCSADLTHAKHYHRRHKVCEFHSKASTVIAAGLTQRFCQQCSR 228
6**********************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.2E-24 | 175 | 228 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 19.956 | 179 | 231 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.92E-22 | 180 | 228 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.9E-17 | 182 | 228 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 232 aa Download sequence Send to blast |
MVDYEWGNPS TILLTGDEAT QEPNQSRQIF DHYTTQEPFS QHSTNNNNYF HQPITNTTLY 60 HHHHQQQQPP PPPPPPHHHH HHNNNNNNSN NAHSLFDPRA YTSASSYAPT HHSMLSIDAA 120 SGGYFMIPKT EEVSRPVDNF ASRIGLNLGG RTYFSSADDD FVSRLYRRPR PGEAGSANIP 180 RCQAEGCSAD LTHAKHYHRR HKVCEFHSKA STVIAAGLTQ RFCQQCSRGL S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 5e-19 | 172 | 228 | 1 | 57 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed during plant development. During pollen development, required for crosporogenesis that leads to archesporial formation and for the histogenesis of the microsporangia. During ovules development, involved in the transition to meiosis of the megaspore mother cells. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:12671094}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in shoot apical region and early floral tissues. Transcripts levels increase in developing pollen sacs, and decrease in later stage of anther development. Strongly expressed in the placental region of the carpels. {ECO:0000269|PubMed:12671094, ECO:0000269|PubMed:17093870}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
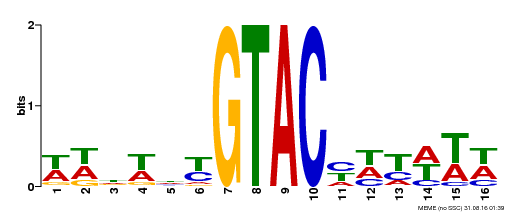
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. Binds specifically to the 5'-GTAC-3' core sequence. Involved in development and floral organogenesis. Required for ovule differentiation, pollen production, filament elongation, seed formation and siliques elongation. Also seems to play a role in the formation of trichomes on sepals. May positively modulate gibberellin (GA) signaling in flower. {ECO:0000269|PubMed:12671094, ECO:0000269|PubMed:16095614, ECO:0000269|PubMed:17093870}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006445348.1 | 1e-171 | squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_006464477.1 | 1e-171 | squamosa promoter-binding-like protein 8 isoform X1 | ||||
| Refseq | XP_024044185.1 | 1e-171 | squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_024949482.1 | 1e-172 | squamosa promoter-binding-like protein 8 isoform X2 | ||||
| Swissprot | Q8GXL3 | 4e-63 | SPL8_ARATH; Squamosa promoter-binding-like protein 8 | ||||
| TrEMBL | A0A067H0X2 | 1e-172 | A0A067H0X2_CITSI; Uncharacterized protein | ||||
| STRING | XP_006464477.1 | 1e-171 | (Citrus sinensis) | ||||
| STRING | XP_006445348.1 | 1e-171 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.2 | 7e-63 | squamosa promoter binding protein-like 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g026888m |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



