 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | orange1.1g044021m | ||||||||
| Common Name | CISIN_1g044021mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 441aa MW: 49911.1 Da PI: 7.7138 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.6 | 5.8e-18 | 185 | 231 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ed l+++v+q+G++ W+ Ia+ m gR +kqc++rw+++l
orange1.1g044021m 185 KGQWTPQEDRMLIRLVAQHGTKKWSVIAKAMT-GRVGKQCRERWYNHL 231
799****************************9.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.2 | 5.8e-16 | 237 | 279 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +W++eEd +l++a+k+ G++ W+ Iar++ gRt++ +k++w+
orange1.1g044021m 237 KEAWSEEEDMILIQAHKEVGNR-WAEIARRLI-GRTENTIKNHWN 279
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 28.037 | 180 | 235 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.35E-30 | 182 | 278 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.1E-17 | 184 | 233 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-28 | 186 | 239 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.10E-15 | 188 | 231 | No hit | No description |
| Pfam | PF13921 | 4.7E-18 | 188 | 245 | No hit | No description |
| PROSITE profile | PS51294 | 20.269 | 236 | 286 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-15 | 236 | 284 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.22E-12 | 239 | 282 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.0E-20 | 240 | 284 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 441 aa Download sequence Send to blast |
MELDSSFRDD NPFISSLFSD NLFKSDLGNG FSSLENPSSK VSPLFQILTI LIDLPSKDPQ 60 KDPPLGISTM SFDPLKGYSN GFLGTESAYT ATPLVLNSTN DVLHGPERRG FWDYSQNFSA 120 HPKPETPNFQ PILMNFQDYE SSSAKLPDEV SCVTGENAYN QDQKRNKRIP VKRERKLPKK 180 NNIIKGQWTP QEDRMLIRLV AQHGTKKWSV IAKAMTGRVG KQCRERWYNH LKPDIKKEAW 240 SEEEDMILIQ AHKEVGNRWA EIARRLIGRT ENTIKNHWNA TKRRQQSKRN TRKQLQENYI 300 KNVTARKNNK SNKGINAAPD QDNSANFDAA VVNADNNMMM MPTSSQSQLG ITSNFASSLV 360 DQWPQVPVYG HHIEAAIGRL SYENGGGYNF GSSVLDEMPS AGSTVESNFE FELALEIDSN 420 YMPEEVKREM ELMEMAYKGL * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 8e-44 | 183 | 285 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 8e-44 | 183 | 285 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 162 | 179 | QKRNKRIPVKRERKLPKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
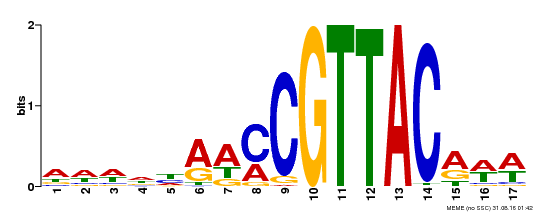
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006446542.1 | 0.0 | transcription factor MYB118 | ||||
| TrEMBL | A0A067EWC6 | 0.0 | A0A067EWC6_CITSI; Uncharacterized protein | ||||
| STRING | XP_006446542.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6052 | 26 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27785.1 | 5e-68 | myb domain protein 118 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | orange1.1g044021m |



