 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | snap_masked-scaffold00829-abinit-gene-0.14-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 309aa MW: 35238.5 Da PI: 4.5476 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 64.3 | 1.7e-20 | 104 | 157 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++++eq++ Le+ Fe ++++ e++++LAkklgL+ rqV vWFqNrRa++k
snap_masked-scaffold00829-abinit-gene-0.14-mRNA-1 104 KKRRLSPEQVHLLEKSFETENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWK 157
56789************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 134.6 | 3.4e-43 | 103 | 193 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlE 60
ekkrrls eqv+lLE+sFe+e+kLeperK++la++LglqprqvavWFqnrRAR+ktkqlE
snap_masked-scaffold00829-abinit-gene-0.14-mRNA-1 103 EKKRRLSPEQVHLLEKSFETENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLE 162
69********************************************************** PP
HD-ZIP_I/II 61 kdyeaLkraydalkeenerLekeveeLreel 91
+dy++Lk++yd+l ++ ++L ke+e+L++e+
snap_masked-scaffold00829-abinit-gene-0.14-mRNA-1 163 RDYDVLKSSYDSLYSNYDTLVKEHEKLKSEM 193
****************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 17.961 | 99 | 159 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.2E-20 | 102 | 163 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 6.42E-20 | 104 | 170 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.22E-19 | 104 | 160 | No hit | No description |
| Pfam | PF00046 | 1.3E-17 | 104 | 157 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-22 | 105 | 166 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.9E-6 | 130 | 139 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 134 | 157 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.9E-6 | 139 | 155 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.0E-13 | 159 | 193 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 309 aa Download sequence Send to blast |
KLCLPTYVLS SFLLSLSLFC VCAPCLVGKT HVSTQDWMME SGRLFFDPTA CRGSNMLFLG 60 NGDNIFRGGR AIMSMEESSK RRSFFSSPDD LFDEEYYDEQ LPEKKRRLSP EQVHLLEKSF 120 ETENKLEPER KTQLAKKLGL QPRQVAVWFQ NRRARWKTKQ LERDYDVLKS SYDSLYSNYD 180 TLVKEHEKLK SEMTIGLQVV SLNEKLQAKG VAGADILAQK AEPLPGDITN VSALQFSVKV 240 EDRLSCGSDV VDEDGPQILD TGDSYFPSDN YPAEDDGSDD GHSYFRNVFV AAEPQHQEDE 300 EPLEWLEWS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 151 | 159 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
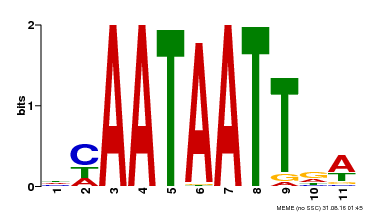
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF529963 | 2e-78 | KF529963.1 Malus domestica homeobox-leucine zipper protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023929344.1 | 1e-179 | homeobox-leucine zipper protein HAT5 isoform X1 | ||||
| Swissprot | Q02283 | 3e-93 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A2I4EUP8 | 1e-141 | A0A2I4EUP8_JUGRE; homeobox-leucine zipper protein HAT5 | ||||
| STRING | EOY02359 | 1e-133 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11283 | 30 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 5e-84 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



